survHE: Survival analysis in health economic evaluation
Code (survHE) Go to project site Code (survHEhmc) Go to project site Code (survHEinla) Go to project site
Survival analysis in health economic evaluation
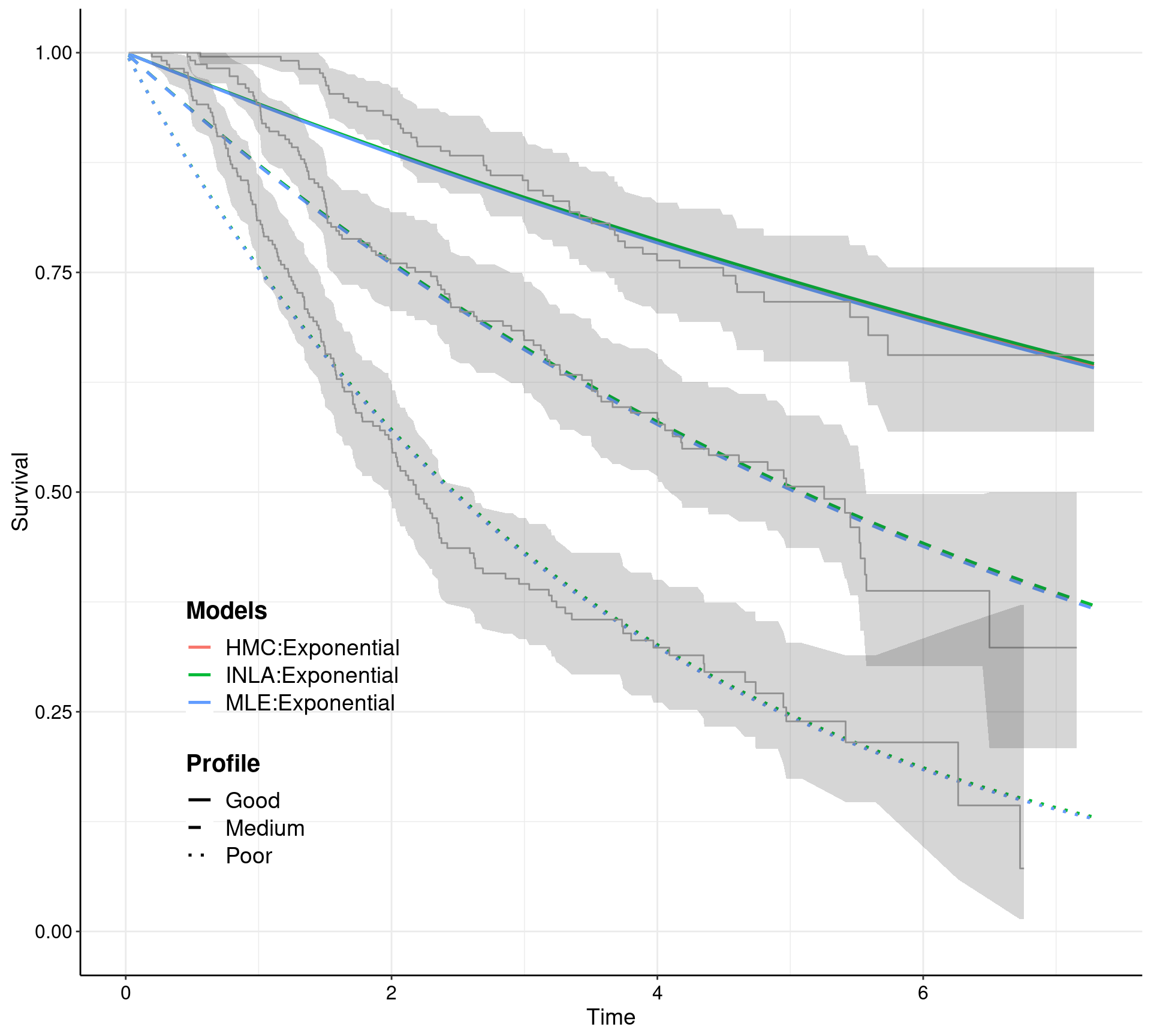
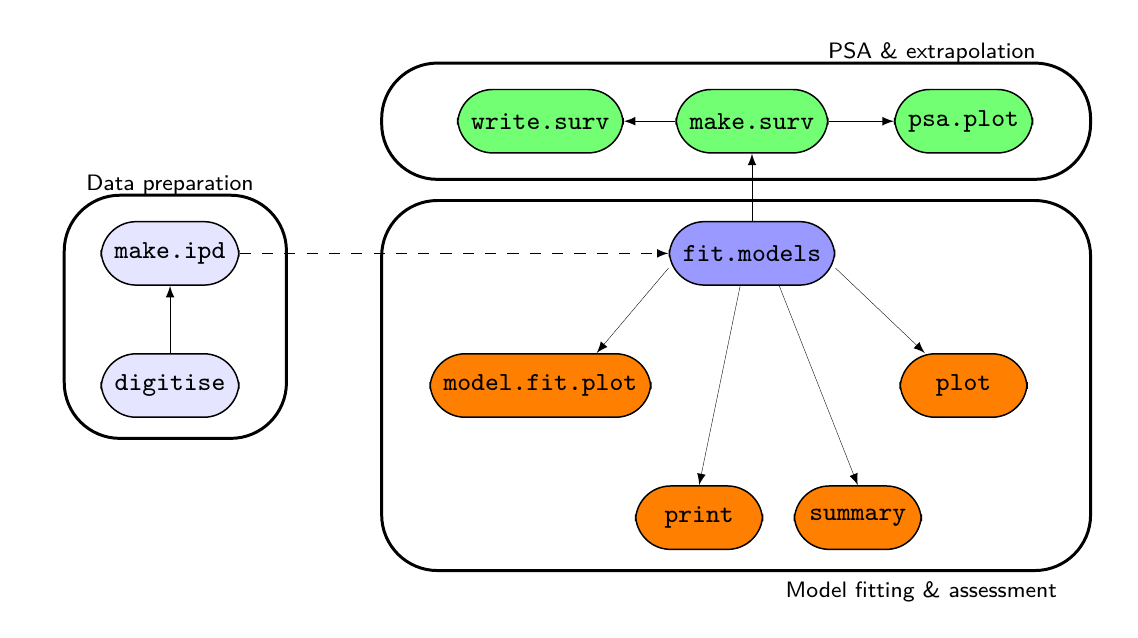
Contains a suite of functions to streamline systematically the workflow involving survival analysis in health economic evaluation. survHE can fit a large range of survival models using both a frequentist approach (by calling the R package flexsurv) and a Bayesian perspective. For a selected range of models, both Integrated Nested Laplace Integration (via the R package INLA) and Hamiltonian Monte Carlo (HMC; via the R package rstan) are possible. HMC models are pre-compiled so that they can run in a very efficient and fast way. In addition to model fitting, survHE provides a set of specialised functions, for example to perform Probabilistic Sensitivity Analysis, export the results of the modelling to a spreadsheet, plotting survival curves and uncertainty around the mean estimates.
survHE can take care of the following modelling aspects:
- Reconstruct individual level dataset from digitised data (e.g. from Kaplan-Meier curves)
- Analyse datasets using a hybrid of R
formulaand specialised commands, i.e.fit.models, which allow the user to select the inferential engine required (mle,inlaorhmc), for a range of parametric models (as suggested e.g. by NICE guidelines) - Perform Probabilistic Sensitivity Analysis directly on the computed parametric survival curves
- Export the output of the statistical model to e.g. a spreadsheet, to complete the economic evaluation (e.g. using Markov models) — of course this step is not necessary and the whole analysis can be embedded in a much bigger (Bayesian) model and performed directly in R!
survHE functions and how they interact
A full documentation (published in the Journal of Statistical Software) is available here.
Installation
The main module (survHE) can be installed directly from CRAN, running the following commands
install.packages("survHE")This installs on your computer the backbone of the package, but, by default, only allows to do the simplest MLE-based model fitting (using flexsurv as the underlying inferential engine). It also installs all the modular facilities to pre- and post-processing the output of the model fitting (including plots and summary tables, as well as the probabilistic sensitivity analysis). The basic survHE module is also available from its GitHub repository; intermediate developments (which can be triggered by pull requests or issues) can be installed using the following commands:
install.packages(
"survHE",
repos = c("https://giabaio.r-universe.dev", "https://cloud.r-project.org")
)In order to “unlock” the Bayesian modelling modules, they need to be installed separately (see also here — although the methods of installation described in that blog post are now superseded: in previous versions of survHE, the two Bayesian modules were stored in “branches” of the main GitHub repository. This was somewhat inefficient and so they now have been promoted to their own separate repositories). Hamiltonian Monte Carlo (HMC) through rstan is made available by installing the module survHEhmc from its GitHub repository, using similar commands to above.
# Bayesian models using HMC/Stan
install.packages(
"survHEhmc",
repos = c("https://giabaio.r-universe.dev", "https://cloud.r-project.org"),
dependencies=TRUE
)This process can be quite lengthy, if you miss many of the relevant packages. Also, the pre-compiled rstan models do take some time at installation (but this steps produces substantial savings at compilation and running time — this is the main reason why survHEhmc has been separated by the default installation of survHE).
Similarly, the Bayesian module to use Integrated Nested Laplace Approximation through the INLA package can be installed from its own GitHub repository using the following commands.
# Bayesian models using INLA
install.packages(
"survHEinla",
repos = c(
"https://giabaio.r-universe.dev",
"https://cloud.r-project.org",
"https://inla.r-inla-download.org/R/stable"
),
dependencies=TRUE
)This is generally rather quick — the bottle neck here is the installation of INLA. It may be useful to change R’s default options in terms of “timeout” (the time spent on a website attempting to download files before giving up), which can be done using the following command (which increases the default from 1 to 10 minutes).
options(timeout=600)This is because INLA is a big package and, depending on your internet connection, it may take longer to download from its own repository.
Installation issues
Previous versions had some installation issues; in particular, installation of the development version via devtools:install_github() could fail in a MS Windows environment with the following error message:
Error in .shlib_internal(args) : C++14 standard requested but CXX14 is not definedThis was due to known issues (see for example here) with new(er) versions of rstan (which survHE uses for full Bayesian modelling). rstan uses by default version 14 of the C++ compiler, so R needs to know and act accordingly. This can be solved by running the following code
dotR <- file.path(Sys.getenv("HOME"), ".R")
if (!file.exists(dotR))
dir.create(dotR)
M <- file.path(dotR, "Makevars.win")
if (!file.exists(M))
file.create(M)
cat("\nCXX14FLAGS=-O3 -Wno-unused-variable -Wno-unused-function",
"CXX14 = $(BINPREF)g++ -m$(WIN) -std=c++1y",
"CXX11FLAGS=-O3 -Wno-unused-variable -Wno-unused-function",
file = M, sep = "\n", append = TRUE)None of this should affect the current versions of survHE and its Bayesian modules.
Last updated: 18 September 2025