| Comparison | A: No intervention | B: Self-help | C: Individual counselling | D: Group counselling |
|---|---|---|---|---|
| AB | 79 / 702 | 77 / 694 | ||
| 18 / 671 | 21 / 535 | |||
| 8 / 116 | 19 / 149 | |||
| AC | 75 / 731 | 363 / 714 | ||
| 2 / 106 | 9 / 205 | |||
| 58 / 549 | 237 / 1561 | |||
| 0 / 33 | 9 / 48 | |||
| 3 / 100 | 31 / 98 | |||
| 1 / 31 | 26 / 95 | |||
| 6 / 39 | 17 / 77 | |||
| 64 / 642 | 107 / 761 | |||
| 5 / 62 | 8 / 90 | |||
| 20 / 234 | 34 / 237 | |||
| 95 / 1107 | 143 / 1031 | |||
| 15 / 187 | 36 / 504 | |||
| 78 / 584 | 73 / 675 | |||
| 69 / 1177 | 54 / 888 | |||
| ACD | 9 / 140 | 23 / 140 | 10 / 138 | |
| AD | 0 / 20 | 9 / 20 | ||
| BC | 20 / 49 | 16 / 43 | ||
| BCD | 11 / 78 | 12 / 85 | 29 / 170 | |
| BD | 7 / 66 | 32 / 127 | ||
| CD | 12 / 76 | 20 / 74 | ||
| 9 / 55 | 3 / 26 |
5. Network meta analysis
Gianluca Baio
Department of Statistical Science | University College London
https://gianluca.statistica.it
https://egon.stats.ucl.ac.uk/research/statistics-health-economics
https://github.com/giabaio https://github.com/StatisticsHealthEconomics
@gianlubaio@mas.to @gianlubaio
Bayesian modelling for economic evaluation of healthcare interventions
València International Bayesian Analysis Summer School, 7th edition, University of Valencia
10 - 11 July 2024
Check out our departmental podcast “Random Talks” on Soundcloud!
Follow our departmental social media accounts + magazine “Sample Space”
Summary
Motivation for Network Meta Analysis (NMA)
- Example: Smoking cessation
Fixed effects meta-analysis
Random effects meta-analysis
References
- Doing Meta-Analysis in R
- Evidence Synthesis for Decision Making in Healthcare
- Bayesian cost effectiveness with the R package BCEA
- NICE DSU Evidence Synthesis Technical Support Document Series
- Data Analysis Using Regression and Multilevel/Hierarchical Models
Multiparameter evidence synthesis
Introduction
- Unusual for a policy question to be informed by a single study
- Must use all available and relevant evidence
- Multiparameter evidence synthesis
- Learning about more than one quantity from combination of direct and indirect evidence
- Example: Network Meta Analysis (NMA)
Network Meta Analysis
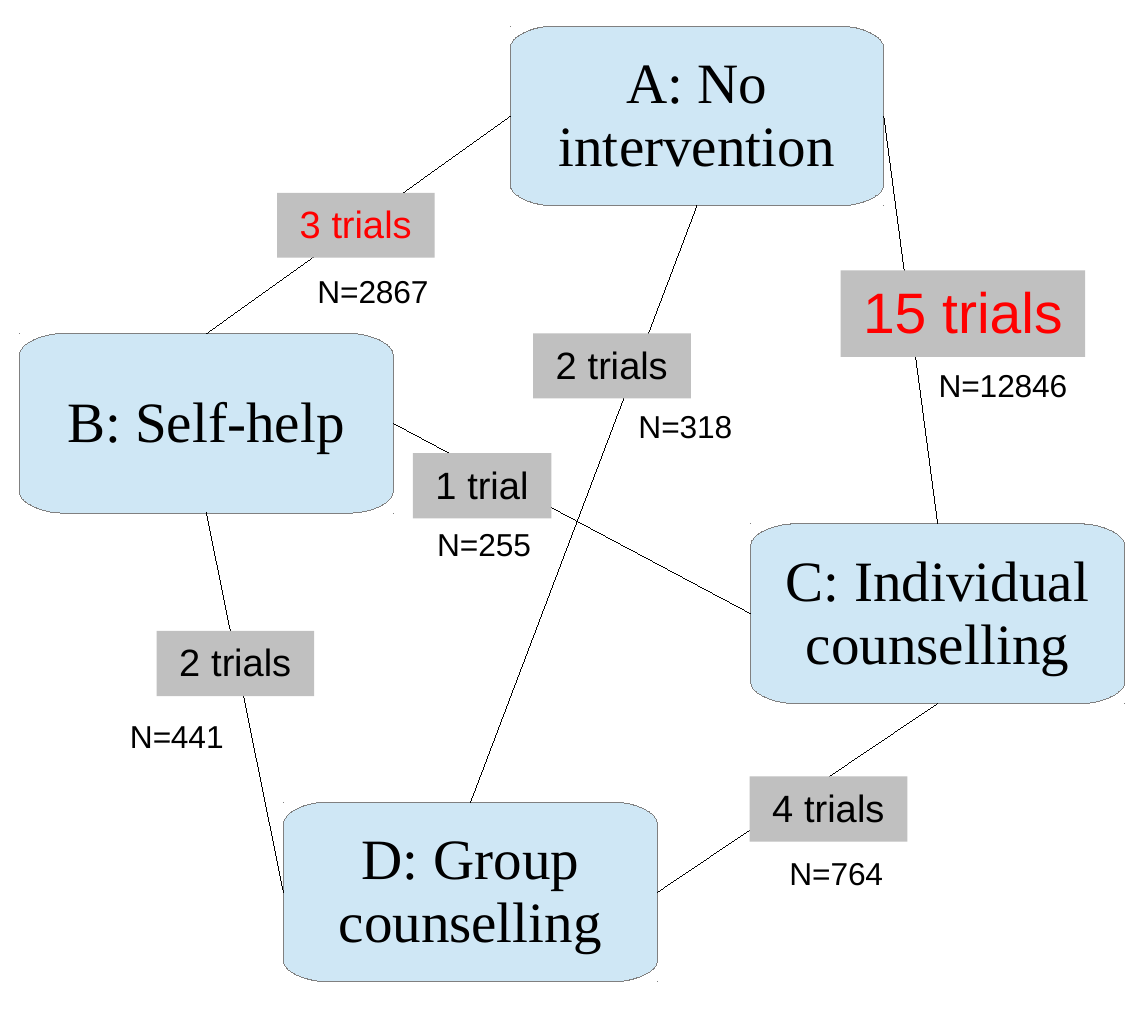
Simplest example
New treatment C: been trialled against old treatment B, but not to A
For health economic evaluation need to compare A/B/C together
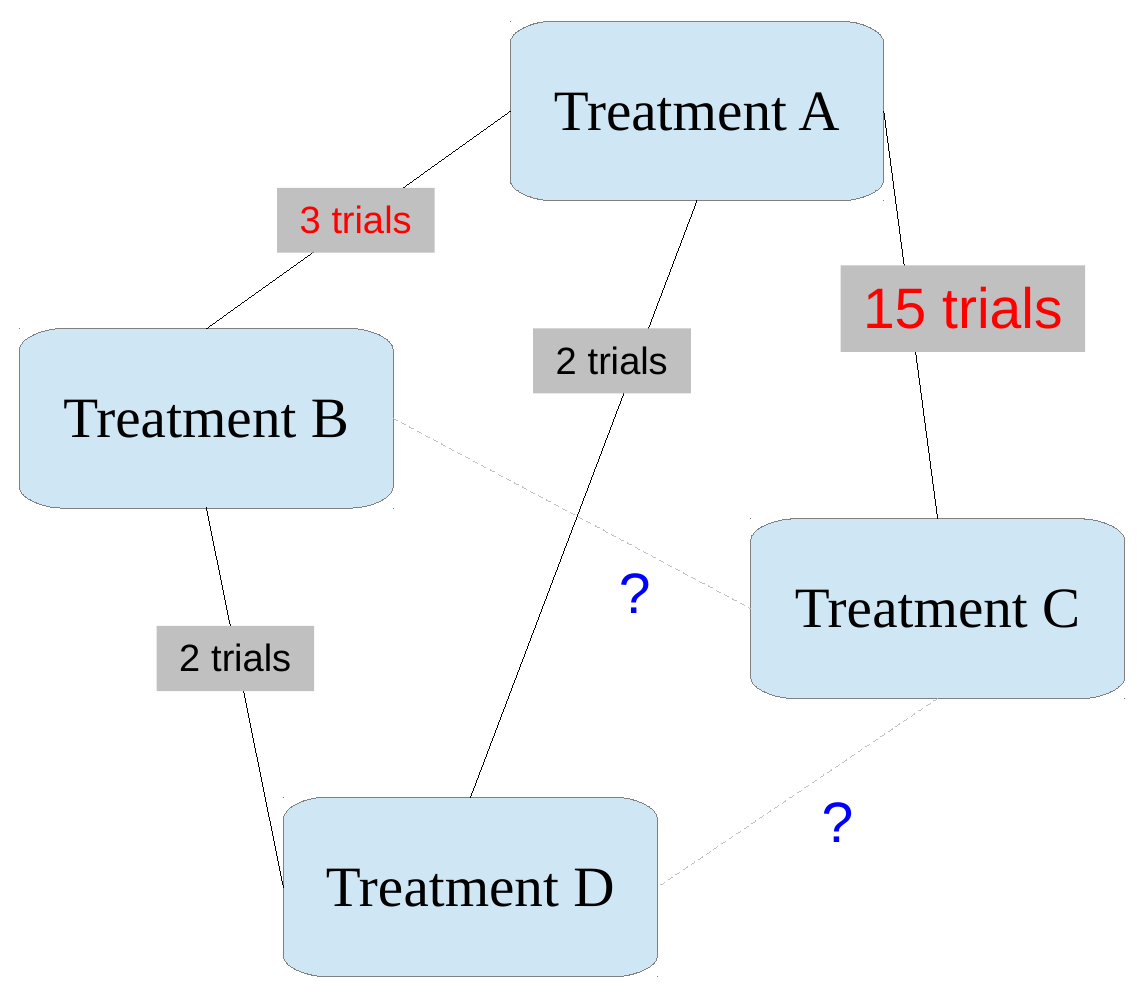
Learn about C/A effect from C/B and B/A trial data
- Also called “mixed treatment comparisons”
- Since can also “mix” direct and indirect data on same comparison…
Common in UK health technology assessment, but require some statistical skills!
Smoking cessation trial
Data
Outcome
Successfully quit smoking by 6-12 months
Number of success / number of participants
Set up
24 trials in total
Network of comparisons involving 4 interventions
Not all interventions tested against all others!
Objective
Estimate the overall effectiveness of the interventions
Potentially add cost-effectiveness analysis
Smoking cessation trial
Network of comparisons
In general…
Network of comparisons
“Fixed effects” NMA
- Log odds of response in each arm modelled as effect of study \(\class{red}{s}\) plus effect of treatment \(\class{red}{t}\) \((s = 1, \ldots , NS\), different values of \(t\) in each \(s)\)
\[\begin{align} & \class{myblue}{r_{st}} \class{myblue}{\sim} \class{myblue}{\dbin(p_{st},n_{st})} \\ & \class{myblue}{\logit(p_{st})} \class{myblue}{=}\class{myblue}{\mu_s + \delta_{st}} \\ & \class{myblue}{\delta_{st}}\class{myblue}{=} \class{myblue}{d_t - d_{t_{s0}}} \end{align}\]
- Study effects \(\mu_s\): log odds in baseline group of study \(s\), considered independent between studies
Treatment effects
\(\delta_{st}\): compared to study \(\class{red}{s}\) baseline \(t_{s_{s0}}\)
\(d_t\): compared to overall baseline treatment \(t=1\) (e.g. placebo) \(\Rightarrow d_1 :=0\) - This essentially means that the effect of treatment \(t=1\) versus the effect of the baseline treatment (again \(t=1\)) is… nothing \((=0)\)!
- “Fixed” effects: \(d_t\) are identical in each study \(s\)
Estimating effects of indirect comparisons
Smoking cessation example
- \(\log \OR\)s \(d_B\), \(d_C\), \(d_D\) (compared to “baseline” treatment A) are directly identifiable from A-B, A-C, A-D trials
But: can deduce indirect comparisons from these basic parameters (with assumptions…)
- \(\log\OR\) of C compared to B is \(d_C-d_B\)
- \(\log\OR\) of D compared to B is \(d_D-d_B\)
- \(\log\OR\) of D compared to D is \(d_D-d_C\)
NB This assumes consistency between indirect and (potential) direct evidence!
- Consider \(t=B\)
- By definition: \(\logit(p_{st}) = \log\left(\frac{p_{st}}{1-p_{st}}\right) =\log\) odds of the event (quit smoking), if you are in group B
- Similarly, \(\logit(p_{sA}) = \log\left(\frac{p_{sA}}{1-p_{sA}}\right) =\log\) odds of the event (quit smoking), if you are in group A \((\Rightarrow t=1)\)
- By definition: \(\displaystyle \OR_{BA}=\frac{\style{font-family:inherit;}{\text{odds}}_B}{\style{font-family:inherit;}{\text{odds}}_A} \Rightarrow \log\OR_{BA}= \log\ \style{font-family:inherit;}{\text{odds}}_B - \log\ \style{font-family:inherit;}{\text{odds}}_A = \logit(p_{sB})-\logit(p_{sA})\)
- So \[\begin{align} \log\OR_{BA} &= \logit(p_{sB})-\logit(p_{sA}) \\ &= \left[\mu_s + \delta_{sB} \right] - \left[\mu_s + \delta_{sA} \right] \\ &= \left[\mu_s +(d_B - d_A)\right] -\left[\mu_s +(d_A - d_A)\right] \\ &= d_B -d_A \\ &= d_B \qquad (\style{font-family:inherit;}{\text{because }} d_A=d_1:=0) \end{align}\]
Manipulating data
Nested indices
[,1] [,2] [,3] [,4]
[1,] 79 77 NA NA
[2,] 18 21 NA NA [,1] [,2] [,3] [,4]
[21,] NA 11 12 29
[22,] NA 7 NA 32
[23,] NA NA 12 20
[24,] NA NA 9 3 [,1] [,2] [,3] [,4]
[1,] 702 694 NA NA
[2,] 671 535 NA NA [,1] [,2] [,3] [,4]
[21,] NA 78 85 170
[22,] NA 66 NA 127
[23,] NA NA 76 74
[24,] NA NA 55 26Coding NMA in BUGS/JAGS
Just write out the equations-ish… 😉
NB:
t[s,a]indicates the treatment associated with studysand its armaVague priors for effects / baseline are typically OK
- But not when the number of comparisons is very small!
for(s in 1:NS) {
for (a in 1:na[s]) {
r[s,t[s,a]] ~ dbin(p[s,t[s,a]], n[s,t[s,a]])
logit(p[s,t[s,a]]) <- mu[s] + delta[s,t[s,a]]
}
# delta are effects compared to arm 1 of each study s
delta[s,t[s,1]] <- 0
for (a in 2:na[s]) {
delta[s,t[s,a]] <- d[t[s,a]] - d[t[s,1]]
}
}
for (i in 1:NS){
# vague prior for baseline log-odds
mu[i] ~ dnorm(0,0.0001)
}
# effect compared to treatment 1 (e.g. placebo)
d[1] <- 0
# vague prior
for (i in 2:NT) {
d[i] ~ dnorm(0, 0.0001)
} Presenting treatment effects
For each treatment \(2, \ldots, NT\) compared to treatment 1 (the reference/baseline: eg “no intervention”/“status quo”, or placebo), can back-transform the \(\log\OR\)s
Results
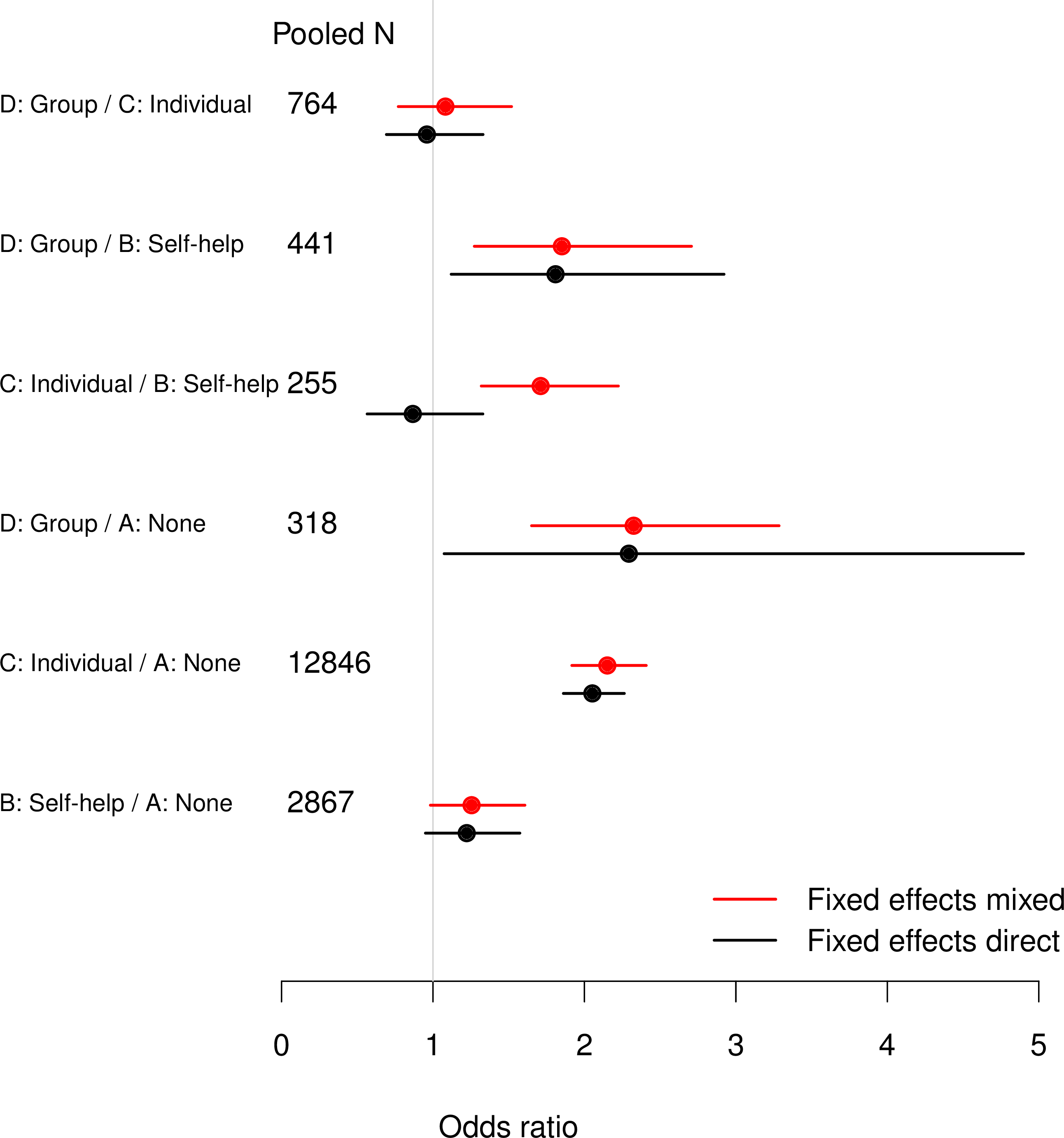
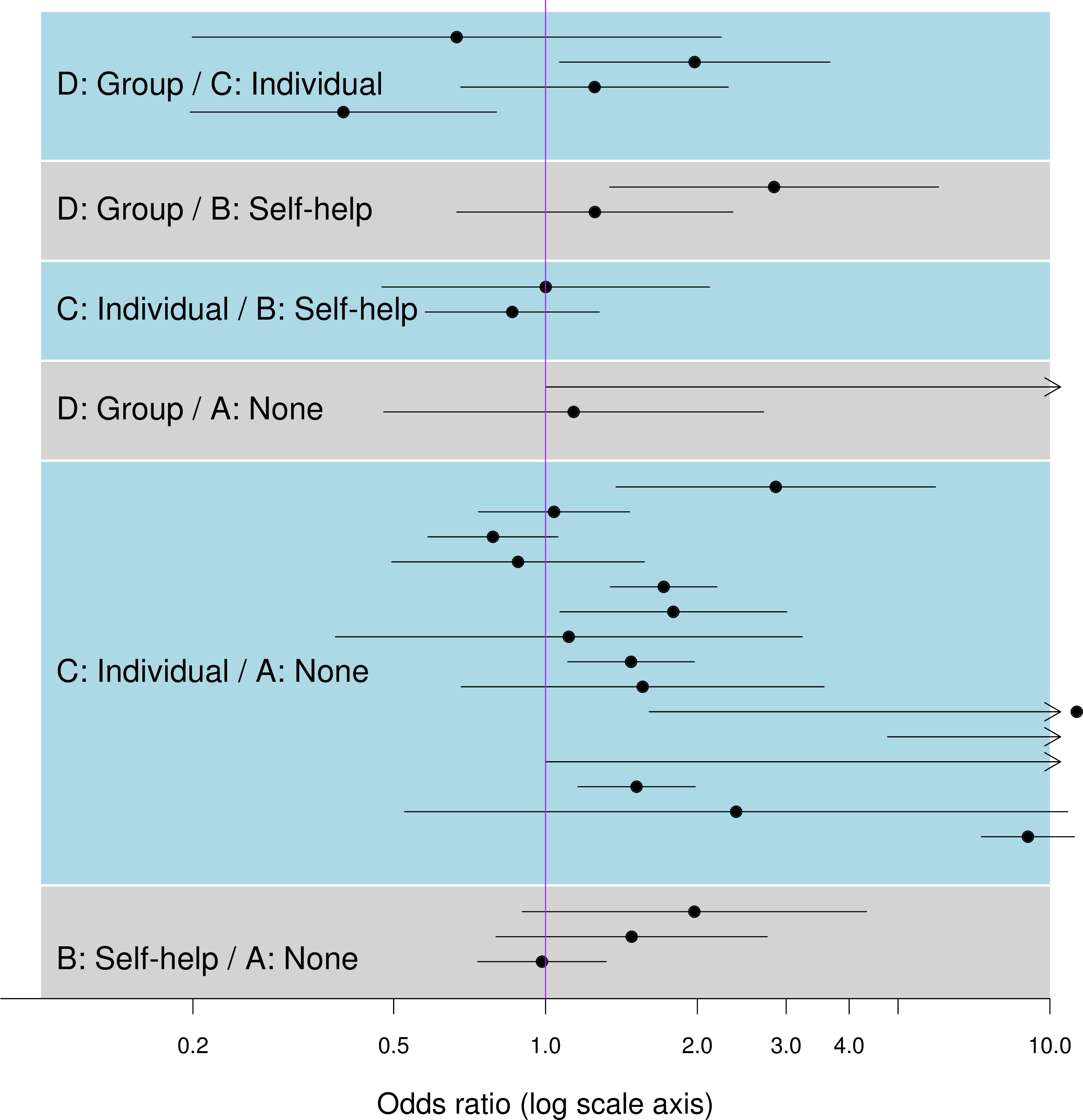
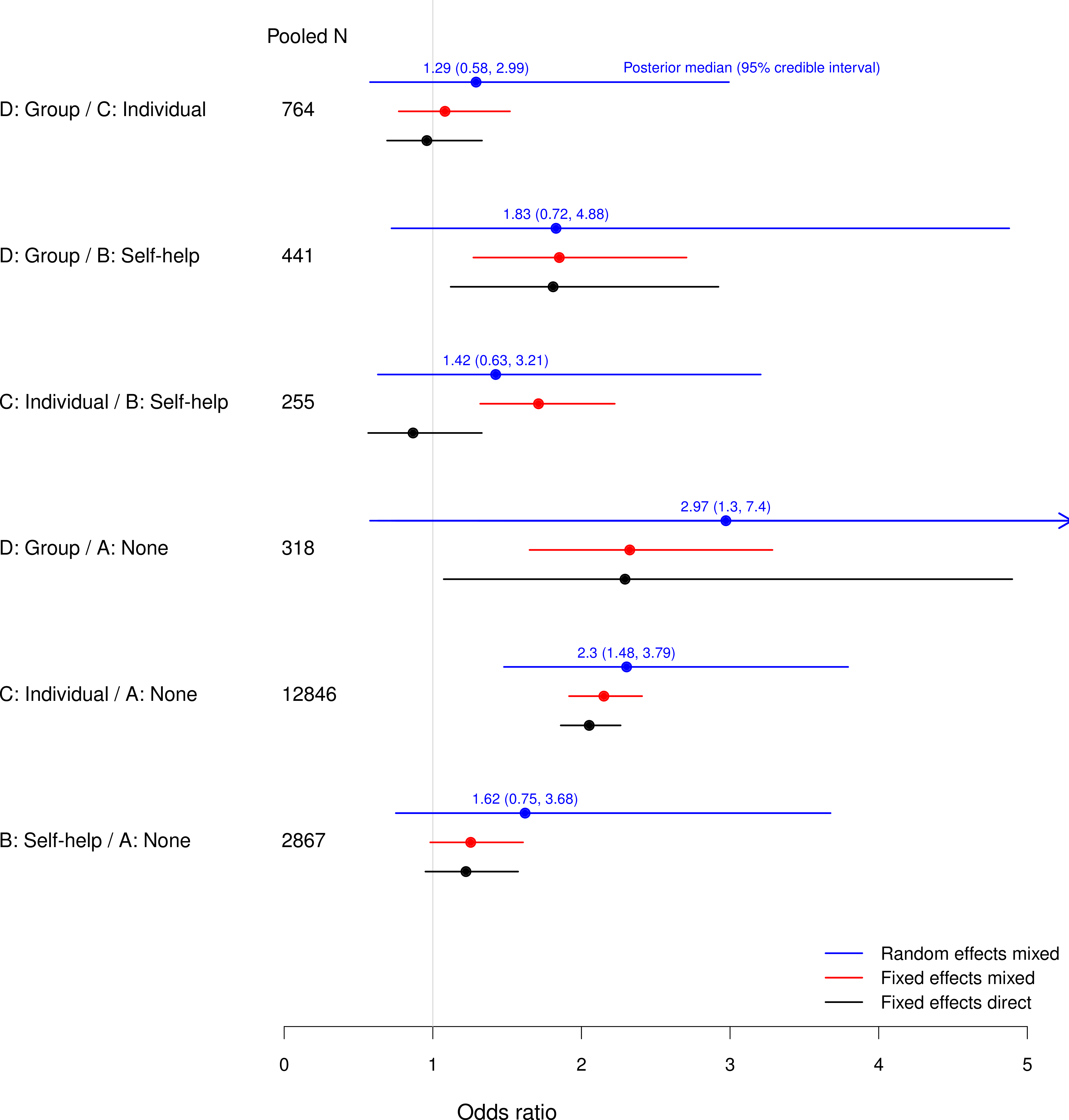
Comparing direct and mixed evidence
Direct-only odds ratios (CIs) from classical analysis of pooled individual data
Results
Heterogeneity between individual studies
Random effects NMA
Replace fixed effects \(\delta_{st}\) of treatment \(t\) in study \(s\)
\[\begin{align} & \class{myblue}{r_{st}} \class{myblue}{\sim} \class{myblue}{\dbin(p_{st},n_{st})} \\ & \class{myblue}{\logit(p_{st})} \class{myblue}{=}\class{myblue}{\mu_s + \delta_{st}} \\ & \class{myblue}{\delta_{st}}\class{myblue}{=} \class{myblue}{d_t - d_{t_{s0}}} \end{align}\]
with a .red[random effect] varying between studies \(s\) with a Normal distribution with mean defined by the fixed effect \[\begin{align} & \class{myblue}{r_{st}} \class{myblue}{\sim} \class{myblue}{\dbin(p_{st},n_{st})} \\ & \class{myblue}{\logit(p_{st})} \class{myblue}{=}\class{myblue}{\mu_s + \delta_{st}} \\ & \class{myblue}{\delta_{st}\sim\dnorm(\mu_{st}^\delta, \sigma^2_{st})} \\ & \class{myblue}{\mu_{st}^\delta}\class{myblue}{=} \class{myblue}{d_t - d_{t_{s0}}} \end{align}\]
still with \(\delta_{st}=0\) for \(t=\) baseline arm of \(s\)
Coding this in BUGS/JAGS
Equations translate relatively straight to BUGS model, again:
is replaced by:
But: a couple of complicating features…
Constraints on random effects variances
- In a NMA, we have
- \(NT\) different treatments
- \((NT - 1)\) different pooled effects, relative to treatment 1 (the baseline / reference) Only 1 effect in standard meta-analysis
- \((NT-1)\) different random effects distributions to estimate?
- Not feasible unless many studies of every single treatment
- \(\Rightarrow\) identifiability constraints needed
- Assume same random effects variance for each treatment comparison
- \(\sigma_{st}^2 = \sigma^2\)
- unless expect differing amounts of heterogeneity for different treatment effects ( Lu and Ades, 2004 and Lu and Ades, 2006)
Prior for \(\class{red}{\sigma^2}\): Uniform from 0 to a large upper limit (eg 10 if on the log scale) is often used, especially to align with standard meta-analysis
- But: Beware of sensitivity to this — particularly if only few studies are considered…
Results
Random effects models
Use in cost-effectiveness analysis
Example
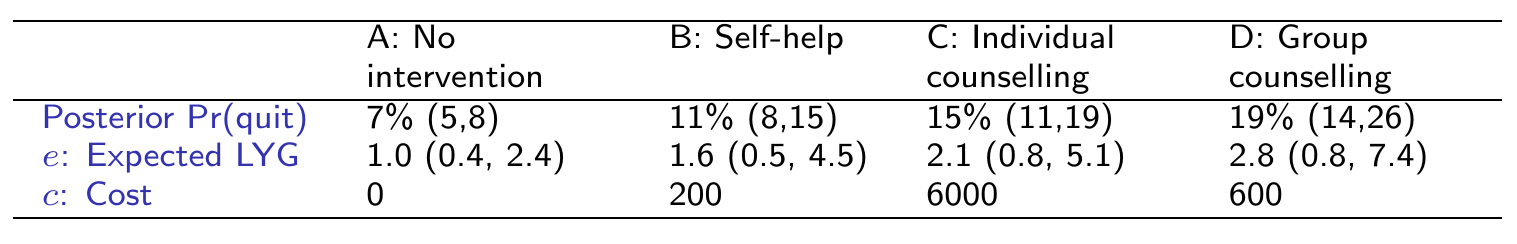
External data on Expected Life-Years Gained if quit smoking:
around 15 years (sd \(\approx\) 4): model as \(L\sim \dnorm(\style{font-family:inherit;}{\text{mean}} =15,\style{font-family:inherit;}{\text{sd}}=4)\)
and code this as
L ~ dnorm(15, 0.0625)inBUGS
Further issues…
Different type of outcomes
- Binary data (Binomial models, as here)
- Counts of events/person-years at risk (Poisson models)
- Mean + sd of continuous outcomes (Normal models) … in each arm of the study
Individual patient data alongside data aggregated by arms
Meta-regression: explain heterogeneity between studies using study-level characteristics as covariates
Detecting / handling conflicts between direct / indirect evidence
1. Intro HTA 2. Bayesian computation 3. ILD 4. ALD 5. NMA © Gianluca Baio (UCL) | | Bayesian models in HTA | VIBASS7 2024 | 10 - 11 July 2024 |